Array Content & Coverage
Explore our protein universe
engines high-content hEXselect and UniPEx protein arrays consist of more than 15,000 human antigens. The broad and unique human proteome coverage gives you the opportunity to analyse interactions with full-length proteins, isoforms, peptides – and even neoantigens such as frameshift peptides as potential biomarkers. Our antigens include major functional classes , cover different tissues and are involved in a broad range of diseases.
Content of engine Protein Arrays
We differentiate the human antigens on our arrays into full-length proteins, protein isoforms & peptides and neoantigens & frameshift peptides. Frameshift mutations are commonly the cause of severe pathologies so neoantigens and frameshift peptides are relevant in different diseases. The combination of established, published and well-characterised antigens and neoantigens enables you to discover hypothesis-free and unbiased novel biomarkers and increases your chance for new, decisive discoveries.
hEXselect Array
The hEXselect Array (product no. 1003) has >57,000 spots and >21,000 human antigens which represents >7,000 different human genes.
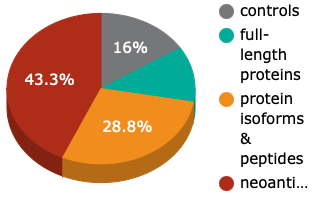
UniPEx Array
The UniPEx Array (product no. 1008) has >36,000 spots and >15,000 human antigens which represents >6,000 different human genes.
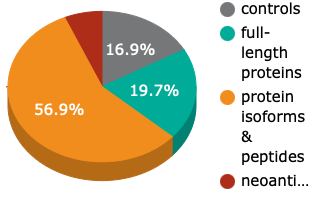
Controls and human protein & peptide expressing E. coli clones are spotted onto a PVDF membrane. Guiding spots, as well as blanks and E. coli clones with empty vectors are used as controls. The E. coli clones are spotted at least in duplicate.
Unique human proteome coverage
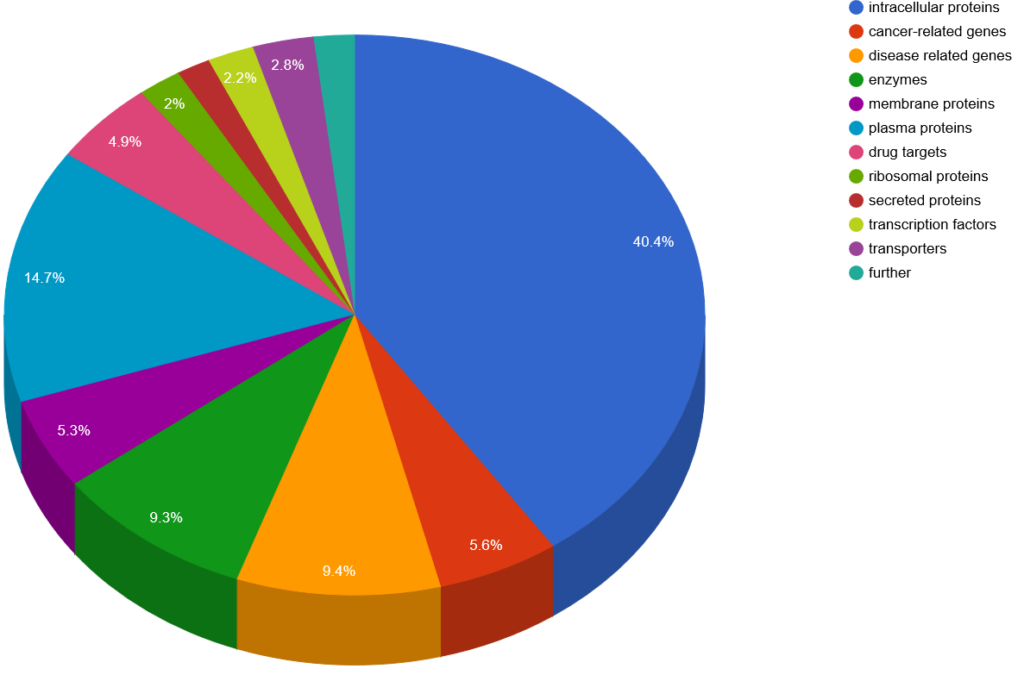
Our antigen library consists of >27,000 different human antigens, represented on hEXselect and/or UniPEx Array. To give you an insight on how these antigens cover the human proteome, we analysed our >2,000 different full-length proteins with the Human Protein Atlas and display in the following charts the general protein classes, tissue expression and disease involvement. Please note that proteins can also be part of several groups, like expression in several tissues.
General protein classes
Protein subclasses of full-length proteins.
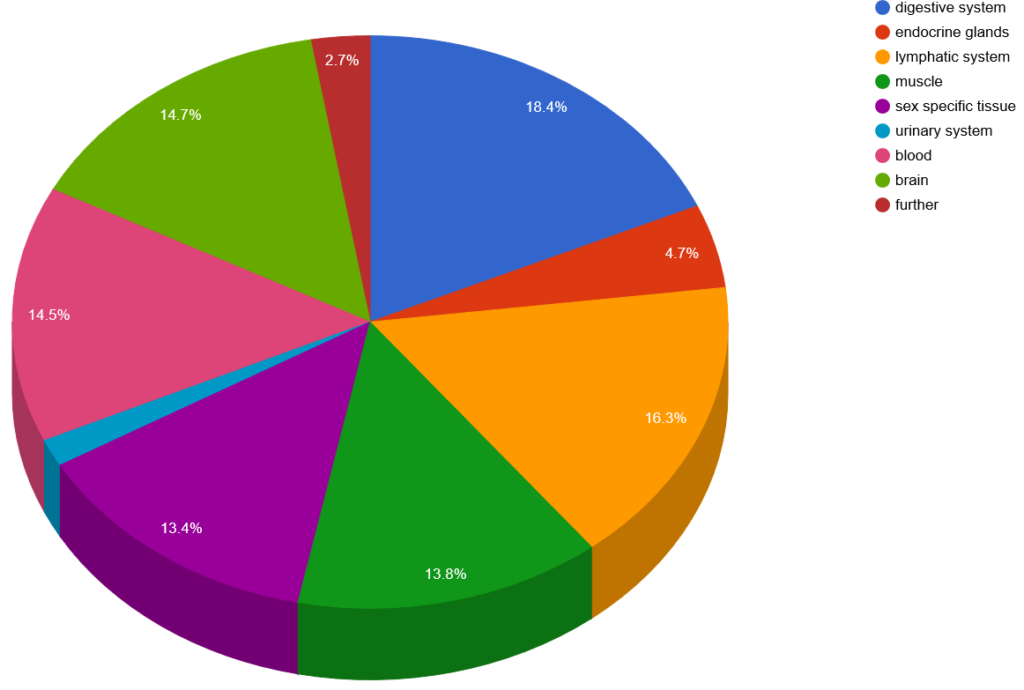
Tissue expression
The human protein expression library is derived from human fetal brain, CD4+ T‑cells, lung and colon cDNA, but our antigens cover a broader range of tissue expression.
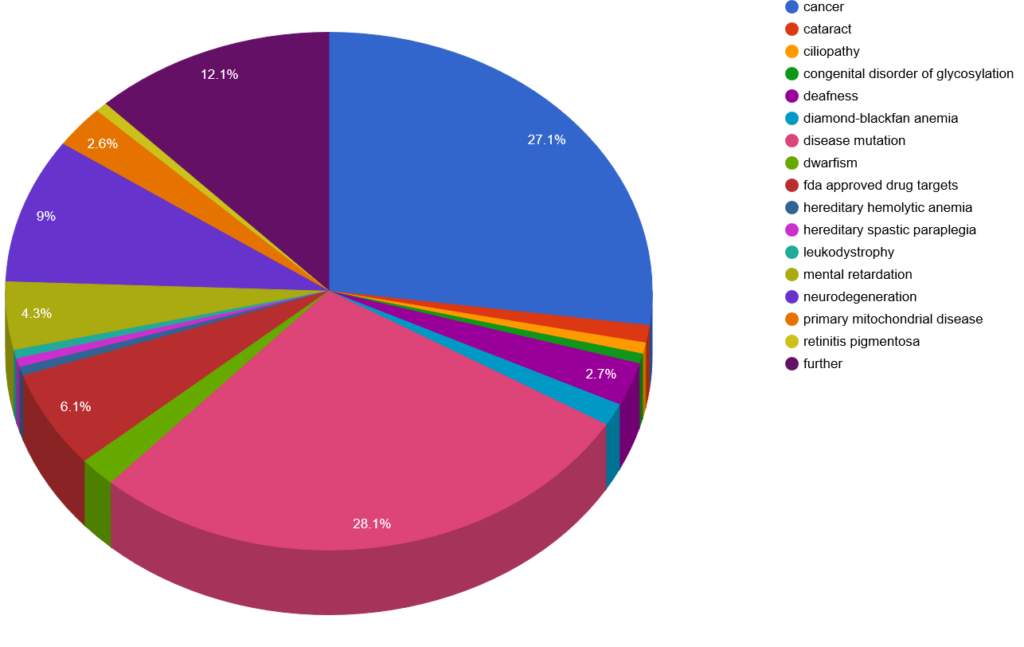
Disease involvement
The full-length proteins are involved in 96 different diseases. With novel biomarker discovery this list will probably expand.
Find your gene of interest!
We have listed in our gene finder database all genes that are on our hEXselect or UniPEx array. Use the full-text search to find the gene from your desired research field.
You need more information?
Download the complete gene finder file (xlsx, 19.3 MB), or just send us a mail.
What’s inside?
- Clone on hEXselect or UniPEx Array
- grouping of full-length, protein isoform or peptide, neoantigen
- name of cloned gene & synonyms based on DNA
- gene ID & HGNC ID of cloned gene
- NCBI URL adress of cloned gene & title of URL adress